Chasing the Ghost Proteome in the Dark matter
Tristan Cardon, Isabelle Fournier, Michel Salzet
Molecular & Cellular Proteomics
https://doi.org/10.1016/j.mcpro.2025.101076
Abstract
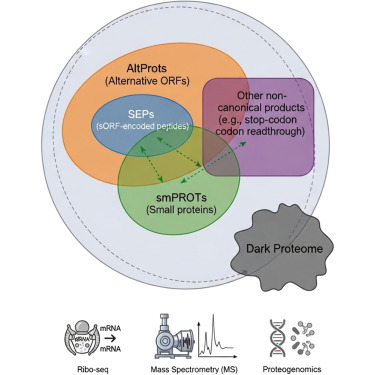
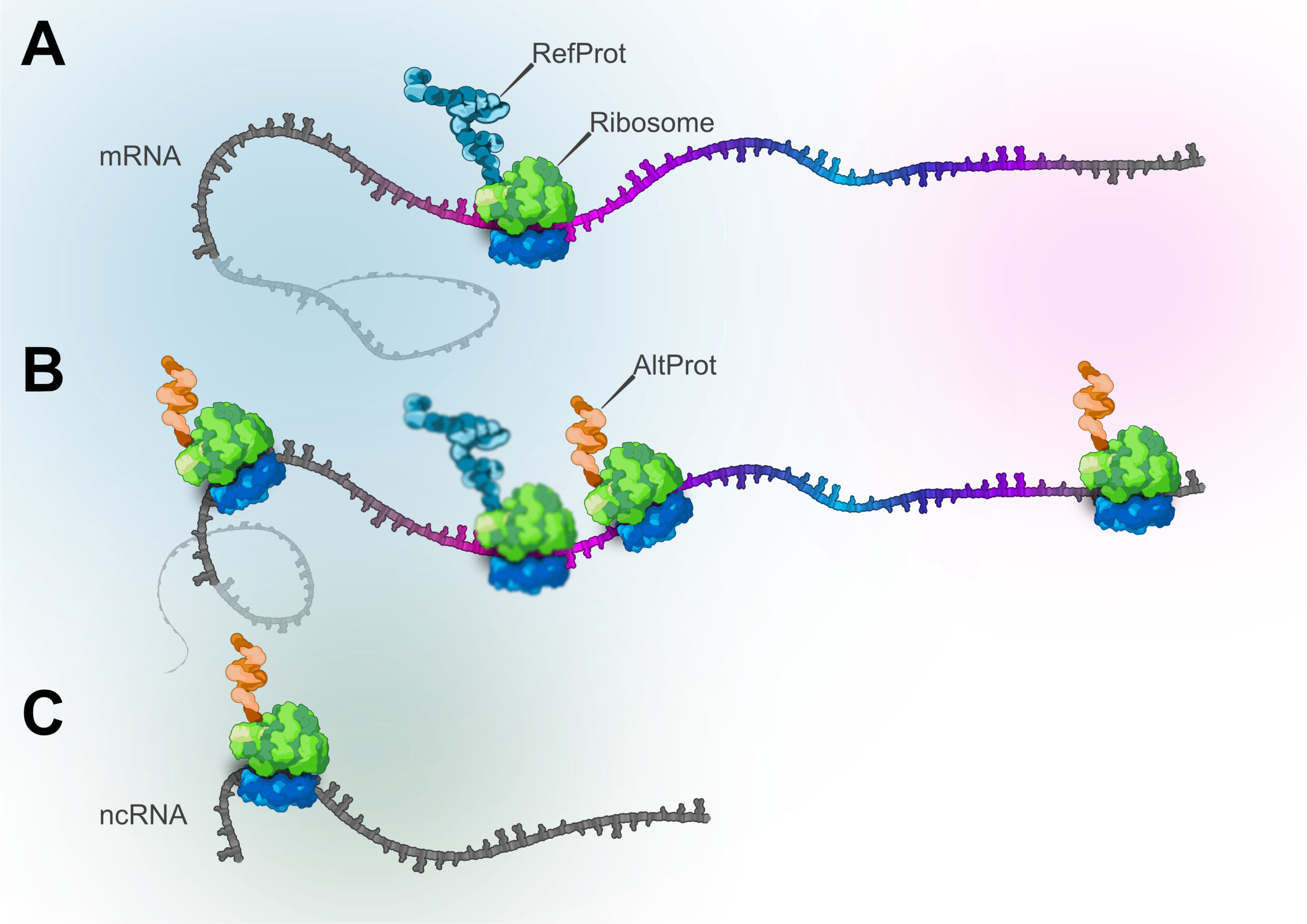
Emerging evidence shows that translation from noncanonical ORFs produces a diverse set of biologically active proteins. These ORFs reside in 5′ and 3′ UTRs, long noncoding RNAs, overlapping frames within annotated genes (dual coding), or pseudogenes and can initiate at non-AUG start codons. The resulting products, variously termed microproteins, small proteins, small ORF–encoded peptides, and alternative proteins, modulate fundamental cellular processes, including metabolic flux and epigenetic regulation. We consolidate these entities under the umbrella of the ghost proteome, a functional proteome arising from the genome’s presumed “dark matter.” This concept is distinct from the dark proteome, which refers to regions of canonical proteins lacking structural, functional, or experimental annotation and is not necessarily derived from noncanonical loci. Recognizing the ghost proteome expands the boundary of what is considered protein coding, demands harmonized nomenclature and database integration, and motivates systematic discovery and functional characterization. By reframing sequences once dismissed as noncoding or “junk,” the ghost proteome compels a re-evaluation of genome annotation and reveals new opportunities to interrogate biology and disease.